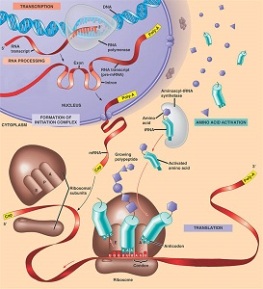
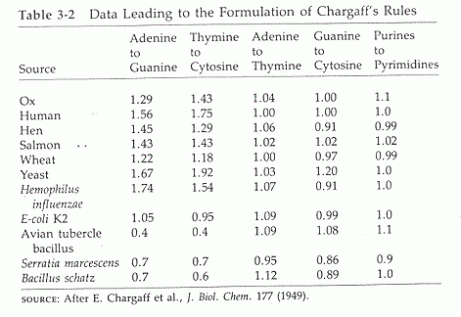
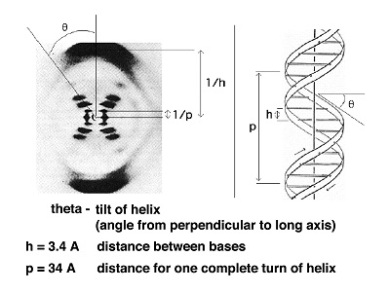
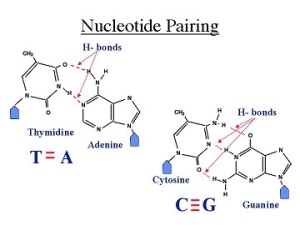
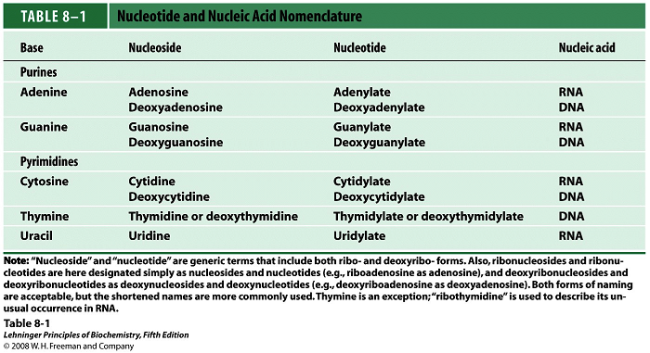
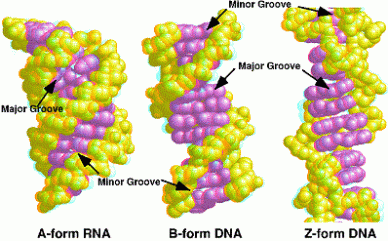
Tca cycle/ krebs cycle/ citric acid cycle……………..
What is it???
Where does is take place???
What is its significance???

Image 98
The pyruvate that was formed in glycolysis goes through oxidation and result in carbon dioxide and water using oxygen. All of this happens in the matrix of the mitochondria and involves the Kreb’s cycle.
First there is a transition stage . In the matrix each pyruvate molecule enters and there is the conversion into an acetyl group. These are carried by coenzyme A as acetylcoenzyme A. There are two carbon atoms in the acetyl groups so when the pyruvate which contains 3 carbons is converted to the acetyl which has two carbons there is the loss of a carbon. This is lost as a result of carbon dioxide in a decarboxylation reaction. There is also a dehydrogenation as there is the reduction of NAD.
STEP 1. There is the combination of the acetic acid subunit of acetyl CoA which has two carbon with a 4 carbon oxaloacetate to form the six carbon compound citrate. After this step due to the process of hydrolysis the coenzyme is released so that it can be able to combine with other acetic acid molecule to start over the Krebs cycle
STEP 2. Isomerization of the citric acid molecule takes place. There is the removal of both a hydroxyl group and a hydrogen molecule and this happen in the form of water. A double bond is formed between the two carbons until there is the replacement of the water molecules. And this result in the formation of isocitrate.
STEP 3. At this stage an NAD molecule oxidizes the isocitrate molecule. The hydrogen atom and the hydroxyl group causes the NAD molecule to become reduced. There is the binding of the NAD molecule and a hydrogen atom. The NAD also takes away the other hydrogen atom leaving behind a carbonyl group. The resulting structure is not stable so there is the release of a carbon dioxide molecule and the resulting structure is a 5 carbon compound known as alpha-ketoglutarate.
STEP 4. Here is where there is the return of the coenzyme A . It comes back to oxidize the alpha-ketoglutarate. There is another reduction of NAD to NADH leaving with another hydrogen. There is instability once again and to restore this a carbonyl group is kicked out as carbon dioxide and there is the replacement of a thioester bond between the previous alpha-ketoglutarate and the coenzyme A creating succinyl coenzyme A complex.
STEP 5. Being generous the water molecules gives its hydrogen atoms to coenzyme A. A greedy free phosphate floating group then pushes out the coenzyme A and this leads to the formation of a bond with the succinyl complex.
STEP 6. Here a molecule of Flavin adenine dinucleotide (FAD) oxidizes succinate. This FAD takes away both of the hydrogen from the succinate forcing a double bond to form between two carbon atoms formingggggg……. fumarate.
STEP 7. Hmmmm To the fumurate an enzyme adds water and this creates malate. This malate is a result of adding to a carbon a hydrogen atom. And then to the carbon that is next to the terminal carbonyl group the addition of a hydroxyl group
STEP 8. Woosh finally the last step…. A NAD molecule oxidizes the malate molecule. The carbon that was initially carrying the hydroxyl group is now turned into a carbonyl group. The final product is the four carbon compound oxaloacetate. This compound can start over the Krebs cycle by combining with acetyl coenzyme A.

Image 99
THE LINK BETWEEN CHEMIOSMOSIS AND THE GENERATION OF ATP IN THE MITOCHONDRIA!!!!!!!
To understand how ATP is generated in the mitochondria by chemiosmosis, we need to break things down first.
Firstly, what is Chemiomosis?????????????????
This is when ions move across a selectively permeable membrane, down an electrochemical gradient. It is paired with ATP generation by moving hydrogen ions across a membrane when cellular respiration is taking place.
CELLULAR RESPIRATION
Cellular respiration involves the breaking down of food (more specifically glucose) to yield energy. This energy is in the form of chemical energy and in the body. The cells of our body recognise this energy in another form known as ATP, so they convert the energy from the food we into ATP which they can use.

Image 100
There are a few terms that we should be familiar with at this stage, but if you’re not, here’s a quick reminder:
- Glucose- this is the main energy source of the body and it is a sugar that consists of 6 carbons.
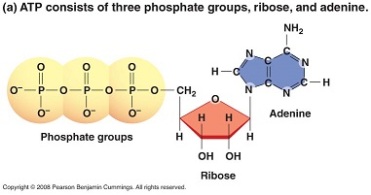
- ATP- (Adenosine Triphosphate) – This is what we would call the main energy or “money source” of the cell. It has a large amount of energy in storage and it also transports that energy in the cell.
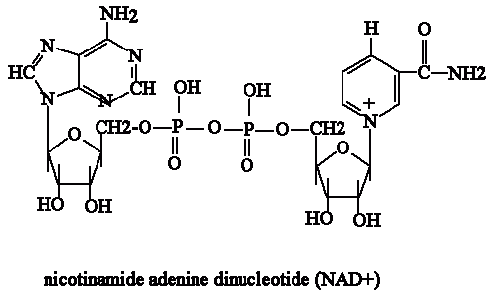
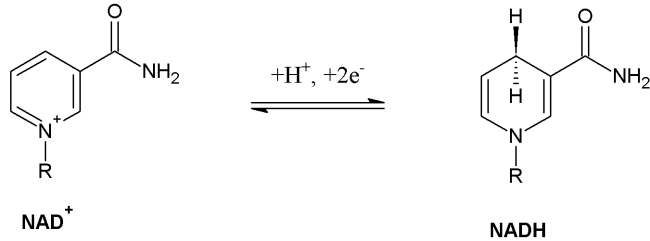
- NADH– An electron carrier with high amounts of energy. It is what transports the electrons made in Glycolysis and Kreb’s cycle to the Electron Transport Chain. In other words, “The Shuttle service.”
- FADH2– Also an electron carrier with a high amount of energy, this is also a “Shuttle Service.”
Cellular respiration takes place in three main steps:
- GLYCOLYSIS
This is when the glucose molecule from food sources is divided into two molecules known as pyruvate. Two ATP molecules are made and also 2 NADH molecules. So for every 1 glucose molecule, we get pyruvate, 2 ATP and 2 NADH.
2. KREB’S CYCLE
Here the pyruvate molecule made in glcolysis is use to 2 ATP and some FADH2 and NADH are also made for the 3rd step.
3. ELECTRON TRANSPORT CHAIN
The NADH and FADH2 made previously are used to form a proton gradient which eventually forms 32 molecules of ATP.
Now that we have recapped what cellular respiration entails, we can now make the link between chemiosmosis and the generation of ATP in the mitochondria.
In the mitochondria of cells, there is something we would call a proton gradient, which is the inner membrane of the mitochondria.
Electrons are journeying through the electron transport chain, hydrogen ions are pushed or pumped out from the mitochondria into a space known as the inter-membrane space.
When this happens, a concentration gradient is formed or a more in depth term would be “proton motive force.”
Proton motive force is simply the energy that causes the movement of protons or the “driving force” of protons.
This causes the protons to move from an area of high concentration to one of lower concentration by diffusion, which in turn, causes the ion to be diffused back into the cell by a process called ATP synthase.
ATP synthase is a form of enzyme that produces ATP by chemiosmosis and it is what allows the protons to be able to go through the membrane.
So to recap: ATP is made in cells that respire by an electrochemical gradient that is found across the inner membrane of the mitochondria. It uses the NADH and FADH2 as the energy source.
Now that the protons have moved back into the membrane by ATP synthase, this same energy that does this, supplies a sufficient amount of energy for ADP to join together with inorganic phosphate to thus form ATP (oxidative phosphorylation).
This is what it mainly looks like………………………………………………..

Image 101
References for Images:
http://blog.cholesterol-and-health.com/2012/01/we-really-can-make-glucose-from-fatty.html
http://guweb2.gonzaga.edu/faculty/cronk/CHEM440pub/L30-index.cfm
http://apps.cmsfq.edu.ec/biologyexploringlife/text/chapter7/concept7.4.html
http://en.wikipedia.org/wiki/Electron_transport_chain